Phenotype Prediction
Motivation
Numerous phenotypic traits can be accurately inferred from DNA methylation (DNAm) data, including immune cell composition29, sex31, smoking status, and both chronological and biological age through the application of epigenetic clocks. These DNAm-derived predictions can be valuable for enriching or completing sample metadata, especially when certain variables were not directly measured during sample collection. Moreover, they serve as an important quality control tool by helping to identify potential sample mix-ups or data inconsistencies—for instance, mismatches between predicted and recorded sex may indicate errors in sample labeling or processing.
Here we outline methods to predict immune cell composition and sex from DNAm data as part of the DNAmArray pipeline.
Cell counts
The EPIDish package can be used to predict blood cell types. It is a R package that infers the proportions of a priori known cell-types present in a sample representing a mixture of such cell-types. Right now, the package can be used on DNAm data of blood-tissue of any age, from birth to old-age, generic epithelial tissue and breast tissue. The package also provides a function that allows the identification of differentially methylated cell-types and their directionality of change in Epigenome-Wide Association Studies.
After proportions of cell types have been estimated, they can be plotted and inspected.
BloodFrac.m_long <- pivot_longer(as.data.frame(BloodFrac.m), cols = colnames(BloodFrac.m))
BloodFrac.m_long %>%
ggplot(aes(y=name, x=value)) +
geom_boxplot() +
theme_bw() + ylab('')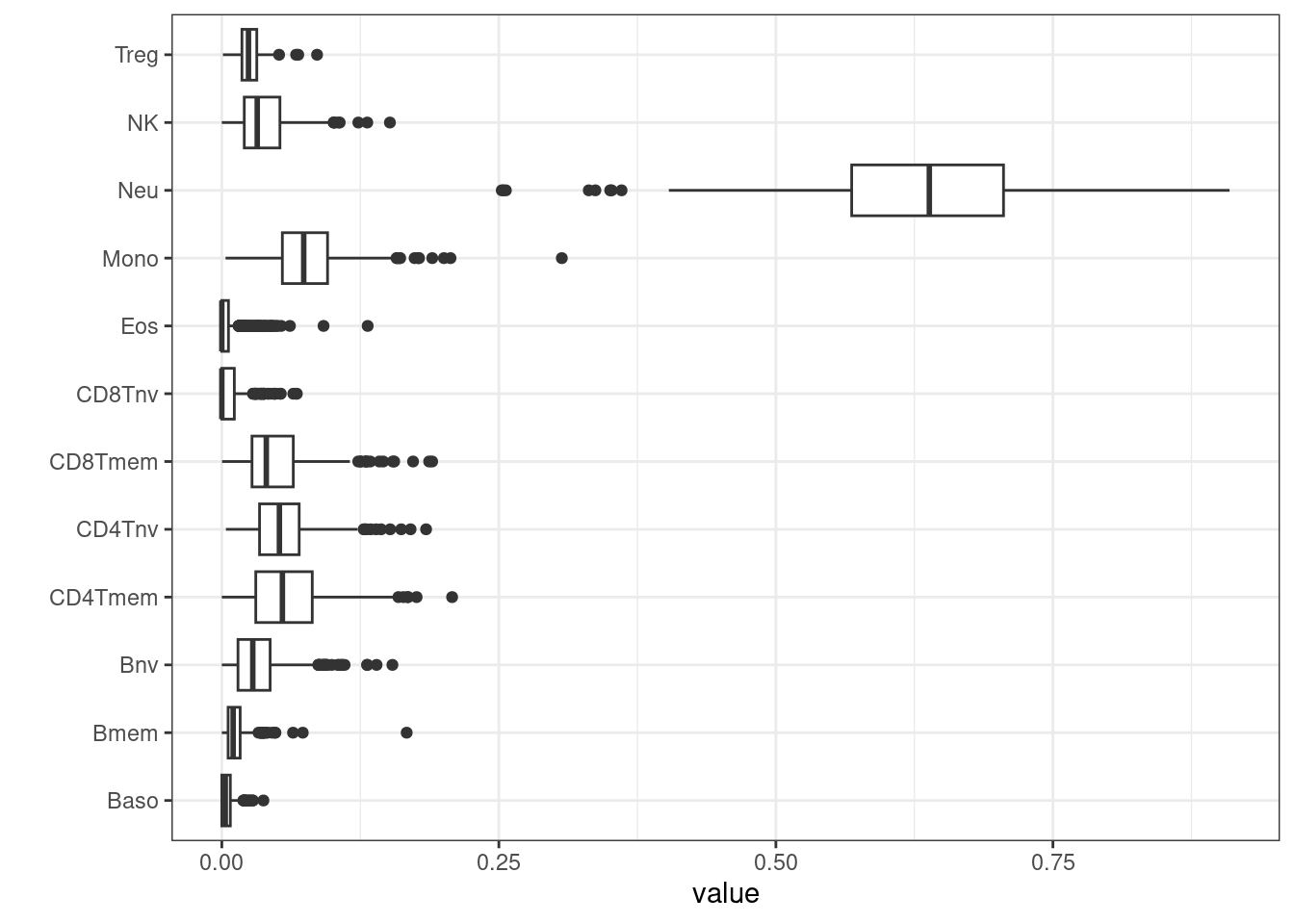
Cell counts can be added to targets for use later when building EWAS models.
##
## TRUE
## 598Other extensions, including UniLIFE which predicts 19 immune cell-types applicable to blood tissue of any age, are available from within EpiDISH, for use in specific contexts29.
Predict Sex
Sex can also be predicted from CpGs on the X-chromosome. Here, we outline the use of estimateSex from wateRmelon31.
## Normalize beta values by Z score...## Fishished Zscore normalization.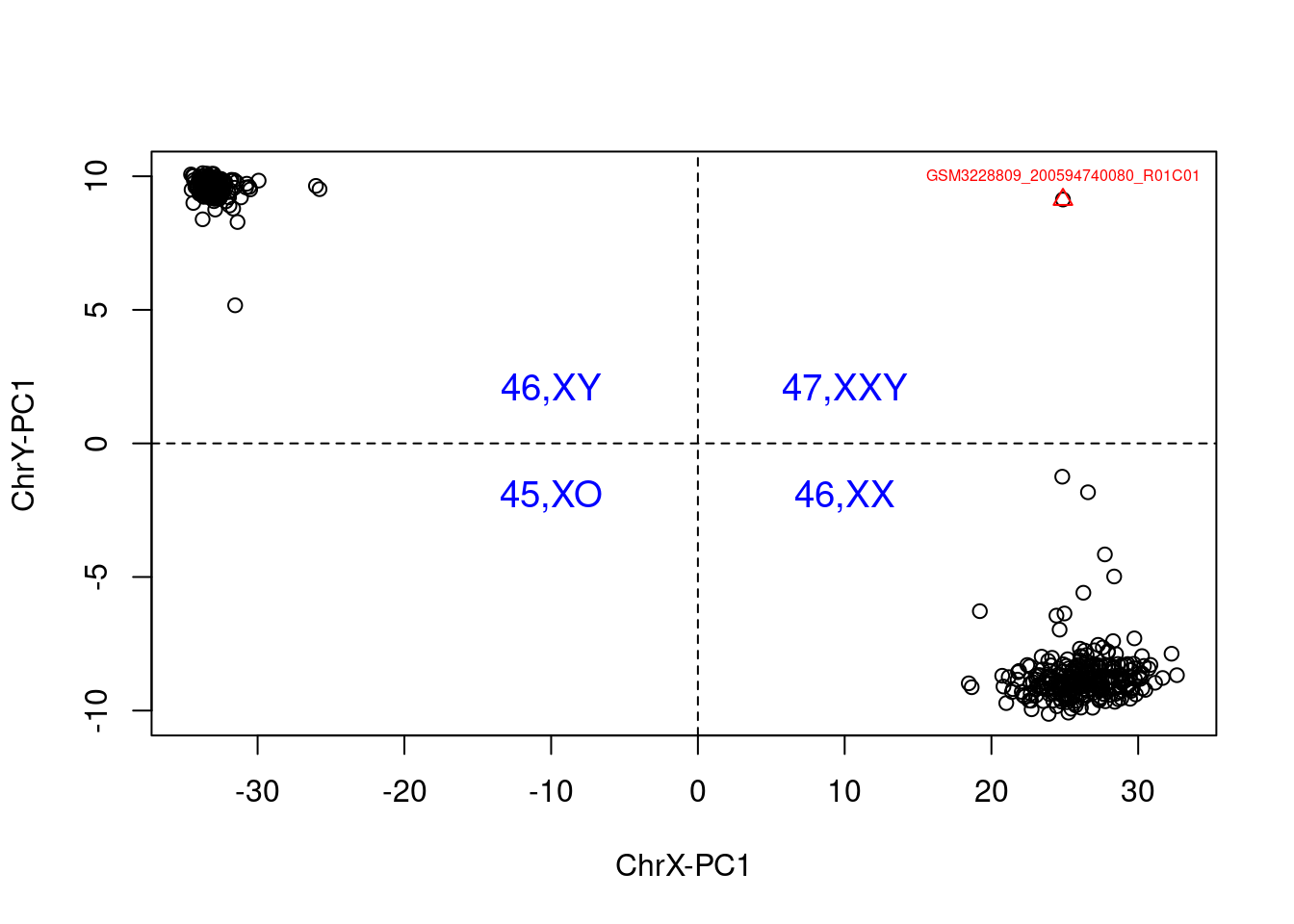
Then a measure can be calculated, determining the sex of each sample and tabulated against recorded sex.
##
## Female Male
## 47,XXY 0 1
## Female 337 1
## Male 2 257As you can see, there is an outlier in the data, which we can remove.
This means that we can feel increased confidence that no incorrect labelling or mix-ups are remaining.